IDPICKER FREE DOWNLOAD
In this diagram, seven peptides observed in human serum are associated with the ceruplasmin sequence from three different species. See the installation instructions for more help. IDPicker assembled protein identifications from the legitimate peptide identifications using parsimony rules. The PSMs that pass the computed probability threshold were considered as confident identifications by PeptideProphet. The same concept was implemented in the new IDPicker by segregating the identifications into separate classes on the basis of peptide NTT and Z state values and calculating their FDRs separately. 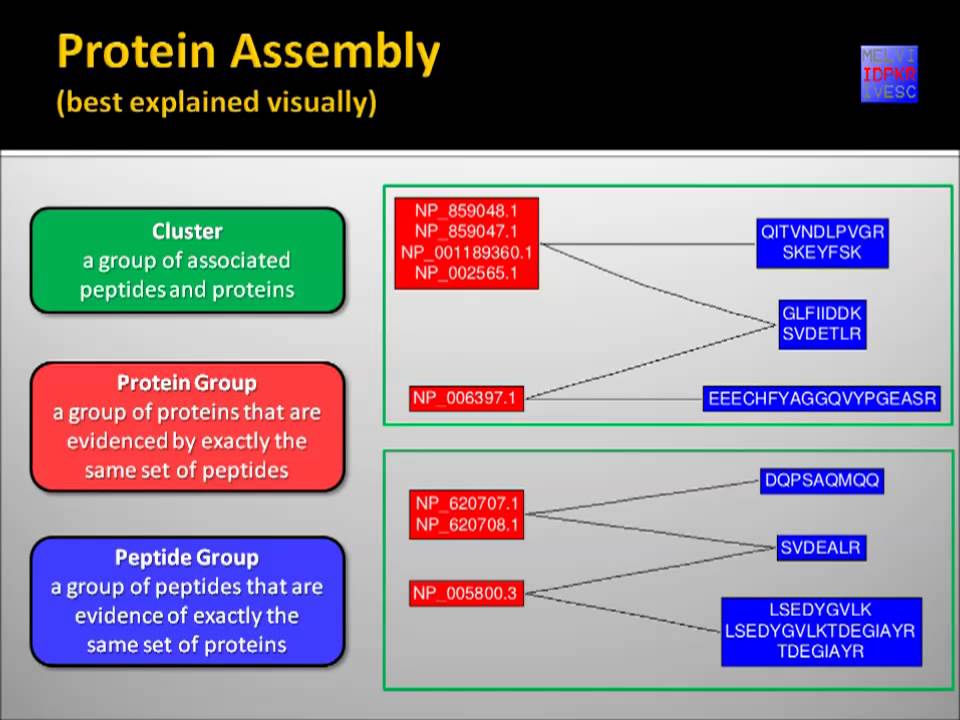
| Uploader: | Grot |
| Date Added: | 6 August 2011 |
| File Size: | 67.44 Mb |
| Operating Systems: | Windows NT/2000/XP/2003/2003/7/8/10 MacOS 10/X |
| Downloads: | 44337 |
| Price: | Free* [*Free Regsitration Required] |
One possible interpretation is that XCorr is very good for ranking peptides for a particular spectrum idpucker is more comparable among multiple spectra when DeltaCN is included. MyriMatch and Mascot were also configured to allow formation of N-terminal pyroglutamate — The resulting peptide mixture was lyophilized and reconstituted with 0. Documentation and example reports are available at http: Assigning significance to peptides identified by tandem mass spectrometry using decoy databases.
IDPicker 2.0: Improved protein assembly with high discrimination peptide identification filtering.
IDPicker has been designed for incorporation in many identification workflows by the addition of a graphical user interface and the ability to read identifications from idipcker pepXML format. Valid peptide identifications were used to assemble a minimal list of protein identifications.
Tandem mass spectra from three different samples were matched to the IPI human protein database version 3.
The new GUI should make the advanced features of the software more accessible to new users. Most search engines report multiple scoring metrics for peptide-spectrum matches.
Many proteomic tools either focus on presenting results for one sample at a time, or struggle with large numbers of samples. These types of searches may idpickeg less sensitive than the concatenated target-decoy database searches used by IDPicker.
Statistical validation of peptide identifications in large-scale proteomics using the target-decoy database search strategy and flexible mixture modeling. Irrespective of the sample type, combining multiple scores from a search engine consistently identified more peptides than using a single score Figure 3. Results and Discussion Combining Multiple Scores Increases Confident Identifications Most search engines report multiple scoring metrics for peptide-spectrum matches.
IDPicker 2.0: Improved Protein Assembly with High Discrimination Peptide Identification Filtering
Footnotes Supporting Information Available: Here, we update IDPicker to increase confident peptide identifications by combining multiple scores produced by database search tools. A peptide that was bounded by two standard trypsin cutting sites yields an NTT of 2. In this diagram, seven peptides observed in human serum are associated with the ceruplasmin sequence from three different species.
We have implemented a new Monte Carlo method in IDPicker that efficiently combines multiple scores, on a per sample basis, using decoy sequences and id;icker discovery rates see Materials and Methods. See the installation instructions for more help.
IDPicker Improved protein assembly with high discrimination peptide identification filtering.
Tandem mass spectra from three different samples were matched to IPI human protein database version 3. Tandem mass spectrometry-based shotgun proteomics has become a widespread technology for analyzing complex protein mixtures. Parsimony Performance Evaluation Data Sets.
Support for reading identifications from pepXML format enables compatibility with any workflow that can produce them. National Center for Biotechnology InformationU. Support Center Support Center. The script was configured to assign a decoy-status of Idlicker for forward hits and R for reverse hits. Survey scans were collected in the Orbitrap at a resolution of 60 within a mass range of — Da. IDPicker has been designed for incorporation in many identification workflows by the addition of a graphical user interface and the ability to read identifications from the pepXML format.
The new IDPicker filter reduces the number of orthologous protein identifications from Swiss-Prot, with minimal effect on the human proteins reported. Mobile phase B consisted of ACN with 0.
IDPicker, however, is able to screen out the mouse and rat sequences by requiring proteins to explain more than one new peptide for inclusion in the final list. Pictoral Guide to IDPicker. Open in a separate window.

Comments
Post a Comment